# Load in required packages, deSolve and socialmixr
# If either package is not installed, install using the install.packages() function
library(deSolve)
library(socialmixr)05b. Using a contact matrix
This is a short guide to how you might use a contact matrix in a model. This uses the socialmixr package to generate a contact matrix from POLYMOD data for the UK, but you can also define your own contact matrices.
First step is to load in the required packages.
We now use socialmixr’s contact_matrix function to build a contact matrix from POLYMOD data for the UK. The age.limits argument below specifies that we will consider 10-year age bands, from 0-9 up to 60-69 and finally 70+.
We then extract the contact matrix itself into mat, and gather some info on the number of groups and their names.
polymod_data <- contact_matrix(polymod, countries = "United Kingdom",
age.limits = seq(0, 70, by = 10))Removing participants that have contacts without age information. To change this behaviour, set the 'missing.contact.age' optionmat <- polymod_data$matrix
n_groups <- nrow(mat)
group_names <- rownames(mat)For more information on socialmixr, try consulting the “socialmixr” vignette by running: vignette("socialmixr", package = "socialmixr").
Let’s take a look at what we have just created:
print(mat) contact.age.group
age.group [0,10) [10,20) [20,30) [30,40) [40,50) [50,60) [60,70)
[0,10) 5.1522843 1.2487310 1.0964467 2.0203046 1.203046 0.6091371 0.3604061
[10,20) 1.2318841 8.0628019 1.1739130 1.7294686 1.710145 0.7004831 0.3719807
[20,30) 1.2711864 1.4406780 3.6016949 1.8898305 1.754237 1.1016949 0.4661017
[30,40) 1.9384615 1.1230769 1.5153846 2.7615385 1.730769 1.2076923 0.6153846
[40,50) 0.7863248 1.8547009 1.6153846 2.1794872 2.897436 1.3504274 0.6837607
[50,60) 0.5583333 0.8916667 1.6333333 1.5500000 1.483333 1.8583333 0.8166667
[60,70) 0.7096774 0.5913978 1.1720430 1.7204301 1.505376 1.4301075 1.1720430
70+ 0.2758621 1.0000000 0.6896552 0.4482759 1.413793 0.9310345 1.2068966
contact.age.group
age.group 70+
[0,10) 0.1421320
[10,20) 0.1545894
[20,30) 0.2033898
[30,40) 0.2923077
[40,50) 0.5982906
[50,60) 0.6333333
[60,70) 0.5483871
70+ 1.3448276print(n_groups)[1] 8print(group_names)[1] "[0,10)" "[10,20)" "[20,30)" "[30,40)" "[40,50)" "[50,60)" "[60,70)"
[8] "70+" Note the usual pattern with a higher number of contacts along the diagonal. Base R’s matrix plotting functions are a bit tricky, but the lattice package has a relatively easy to use function:
lattice::levelplot(mat)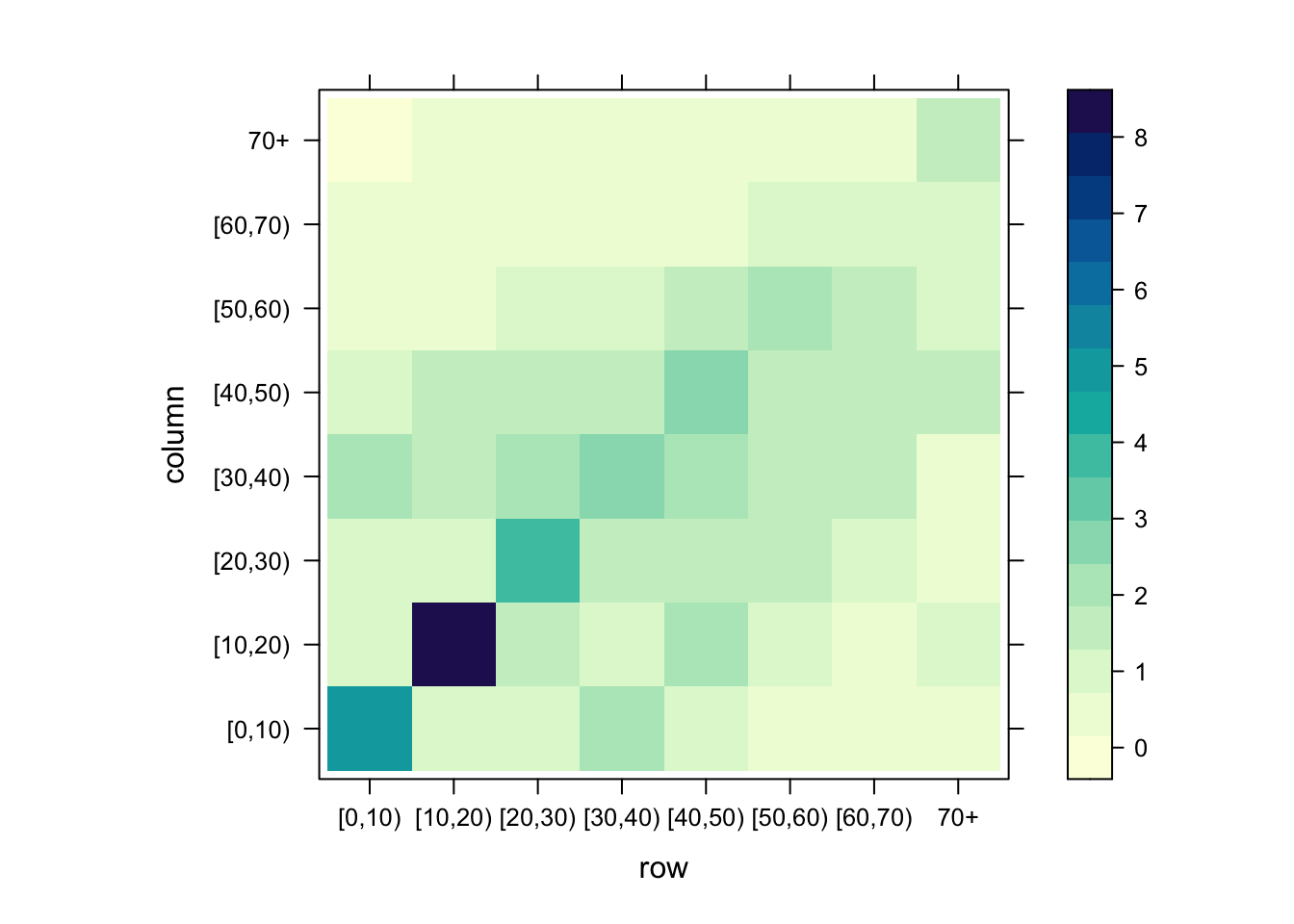
Now let’s start bringing together our model function, initial state, parameters, and times, as normal for an ODE model.
First, the model function:
# Define model function
SIR_conmat_model <- function(times, state, parms)
{
# Get variables
S <- state[1:n_groups]
I <- state[(n_groups + 1):(2 * n_groups)]
R <- state[(2 * n_groups + 1):(3 * n_groups)]
N <- S + I + R
# Extract parameters
p <- parms[["p"]]
cm <- parms[["cm"]]
gamma <- parms[["gamma"]]
# Build force of infection
lambda <- (p * cm %*% (I/N))[,1]
# Define differential equations
dS <- -lambda * S
dI <- lambda * S - gamma * I
dR <- gamma * I
res <- list(c(dS, dI, dR))
return (res)
}There’s a lot to unpack here, so let’s go step by step.
First, our state vector needs to have n_groups versions of the S compartment, n_groups versions of the I compartment, and n_groups versions of the R compartment. Within the vector, we’ll lay it out in this order: \((S_1, S_2, ..., S_n, I_1, I_2, ..., I_n, R_1, R_2, ..., R_n)\). So, in the lines
# Get variables
S <- state[1:n_groups]
I <- state[(n_groups + 1):(2 * n_groups)]
R <- state[(2 * n_groups + 1):(3 * n_groups)]
N <- S + I + Rwe’re extracting S, I, and R each as vectors holding n_groups numbers each. Then we’re defining N as the sum of those vectors; it will also hold n_groups numbers, the first one corresponding to the 0-9 age group, the second one corresponding to the 10-19 age group, and so on.
Next we extract parameters:
# Extract parameters
p <- parms[["p"]]
cm <- parms[["cm"]]
gamma <- parms[["gamma"]]Notice here we are using double brackets [[ instead of single brackets [[ like we normally do. This is because cm will be our contact matrix, and you can’t really store a matrix inside a vector. You can store it in a list though, which takes double brackets to extract elements.
Now we calculate our force of infection.
# Build force of infection
lambda <- (p * cm %*% (I/N))[,1]This is a bit cryptic, so let’s break it down. What we are doing here is calculating \(\lambda_i = \sum_j c_{ij} I_j/N_j\), for all \(i\). One way of doing this would be with a for loop:
lambda = rep(0, n_groups)
for (i in 1:n_groups) {
lambda[i] <- sum(p * cm[i, ] * I/N)
}The line lambda <- (p * cm %*% (I/N))[,1] is equivalent to this, and uses matrix multiplication %*% to achieve what it’s doing. This will run a bit faster in R than the for loop, but either way of calculating the force of infection, lambda (\(\lambda\)), is probably fine.
Finally we define and return the differential equations:
# Define differential equations
dS <- -lambda * S
dI <- lambda * S - gamma * I
dR <- gamma * I
res <- list(c(dS, dI, dR))
return (res)Remember, S, I, and R are vectors, so dS, dI, and dR are also vectors with length n_groups. The line res <- list(c(dS, dI, dR)) sticks them all together into a vector with n_groups\(\,\times 3\) elements. Then this gets returned.
OK, that’s the end of the model function. Carrying on, we set up the parameters and times (remember, parms will be a list this time instead of a vector):
# Define parameters
parms <- list(p = 0.05, cm = mat, gamma = 0.2) # NOT c(p = 0.05, cm = mat, gamma = 0.2) !
# Define time to run model
times <- seq(from = 0, to = 50, by = 1)And define our initial conditions:
# Define initial conditions
N0 <- seq(1000, 650, length.out = n_groups)
I0 <- rep(1, n_groups)
S0 <- N0 - I0
R0 <- rep(0, n_groups)
names(S0) <- paste0("S", 1:n_groups)
names(I0) <- paste0("I", 1:n_groups)
names(R0) <- paste0("R", 1:n_groups)
state <- c(S0, I0, R0)Note that this sets up the population size so there are 1000 0-9 year olds, 650 70+ year olds, and intermediate numbers in between. Let’s look at our initial state to make sure it’s correct:
print(state) S1 S2 S3 S4 S5 S6 S7 S8 I1 I2 I3 I4 I5 I6 I7 I8 R1 R2 R3 R4
999 949 899 849 799 749 699 649 1 1 1 1 1 1 1 1 0 0 0 0
R5 R6 R7 R8
0 0 0 0 Looks good.
Now all we need to do is solve the ODE system and plot it as before:
# Solve equations
output_raw <- ode(y = state,
times = times,
func = SIR_conmat_model,
parms = parms)
# Convert to data frame for easy extraction of columns
output <- as.data.frame(output_raw)
# Plot output
par(mfrow = c(1, 1))
matplot(output$time, output[, 2:ncol(output)], type = "l", xlab = "Time (years)", ylab = "Number of people")
We use matplot here to quickly plot a large number of different lines, each stored in a different column of a matrix. Cool plot, but not so easy to see what’s going on, so let’s split it out by S versus I versus R:
# Make a 2x2 grid
par(mfrow = c(2, 2))
# Plot S, I, R on separate plots
matplot(output$time, output[, (0 * n_groups + 2):(1 * n_groups + 1)], type = "l",
xlab = "Time (years)", ylab = "Susceptible", col = rainbow(n_groups), lty = 1)
matplot(output$time, output[, (1 * n_groups + 2):(2 * n_groups + 1)], type = "l",
xlab = "Time (years)", ylab = "Infectious", col = rainbow(n_groups), lty = 1)
matplot(output$time, output[, (2 * n_groups + 2):(3 * n_groups + 1)], type = "l",
xlab = "Time (years)", ylab = "Recovered", col = rainbow(n_groups), lty = 1)
# Put a legend in the bottom right of the grid
plot.new()
legend("center", legend = rownames(mat), col = rainbow(n_groups), lty = 1)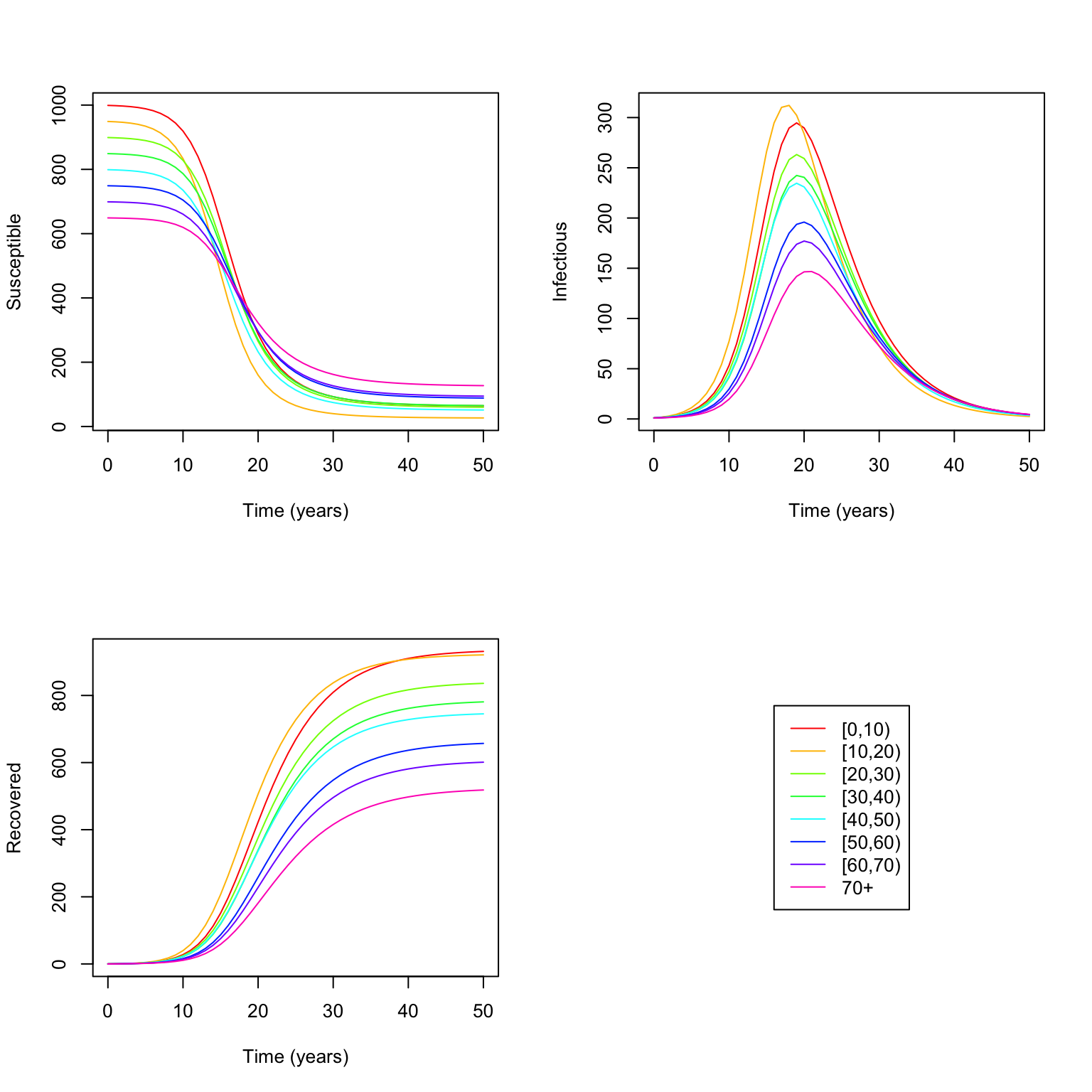
# Put plot grid back to 1x1
par(mfrow = c(1, 1))