Local area reproduction numbers and S-gene target failure
This study has not yet been peer reviewed.
Read the current verision of the work in progress report
Analysis code avilable at https://github.com/epiforecasts/covid19.sgene.utla.rt - contributions welcome!
Background: Local estimates of the time-varying effective reproduction number (Rt) of Covid-19 in England have become increasingly heterogeneous since the introduction of the November national lockdown in England. This variation may be attributed to the spread of a novel Covid-19 variant. In this report we aimed to investigate the changes in the proportion of positive cases that were S-gene negative (“S-gene target failure”, SGTF), an indicator of the novel variant, and to correlate them with changes in the estimated reproduction number at the level of upper tier local authorities (UTLA).
Method: We explored the relationship between the proportion of samples that were S-gene negative and the effective reproduction number of test positive cases over time. Effective reproduction numbers were estimated using the EpiNow2 R package independently for each local authority using two different estimates of the generation time. The brms R package was used to fit a range of regression models to estimate a multiplicative relationship between SGTF and weekly mean R. Models were compared using the expected log-predictive density.
Results: We found evidence of an association between increased mean Rt and the proportion of SGTF across all models evaluated with the magnitude of the effect increasing as model flexibility was decreased. Models found an effect consistent with the low ends of estimates that reported elsewhere when a generation time with a mean of 5.5 days was used. Models that adjusted for either national level or NHS region level residual variation over time were found to fit the data better and found yielded consistent results of the association of Rt with SGTF.
Conclusions: Our results indicate that even after adjusting for between NHS region residual variation over time S-gene negativity was associated with an increase in the effective reproduction number of Covid-19. These findings were robust across a range of models and generation time assumptions though the specific effect size was variable depending on the assumptions used. The lower bound of the estimated effect indicated that NPI measures implemented between September and January 1st in England may not be sufficient to reduce the reproduction number below 1.
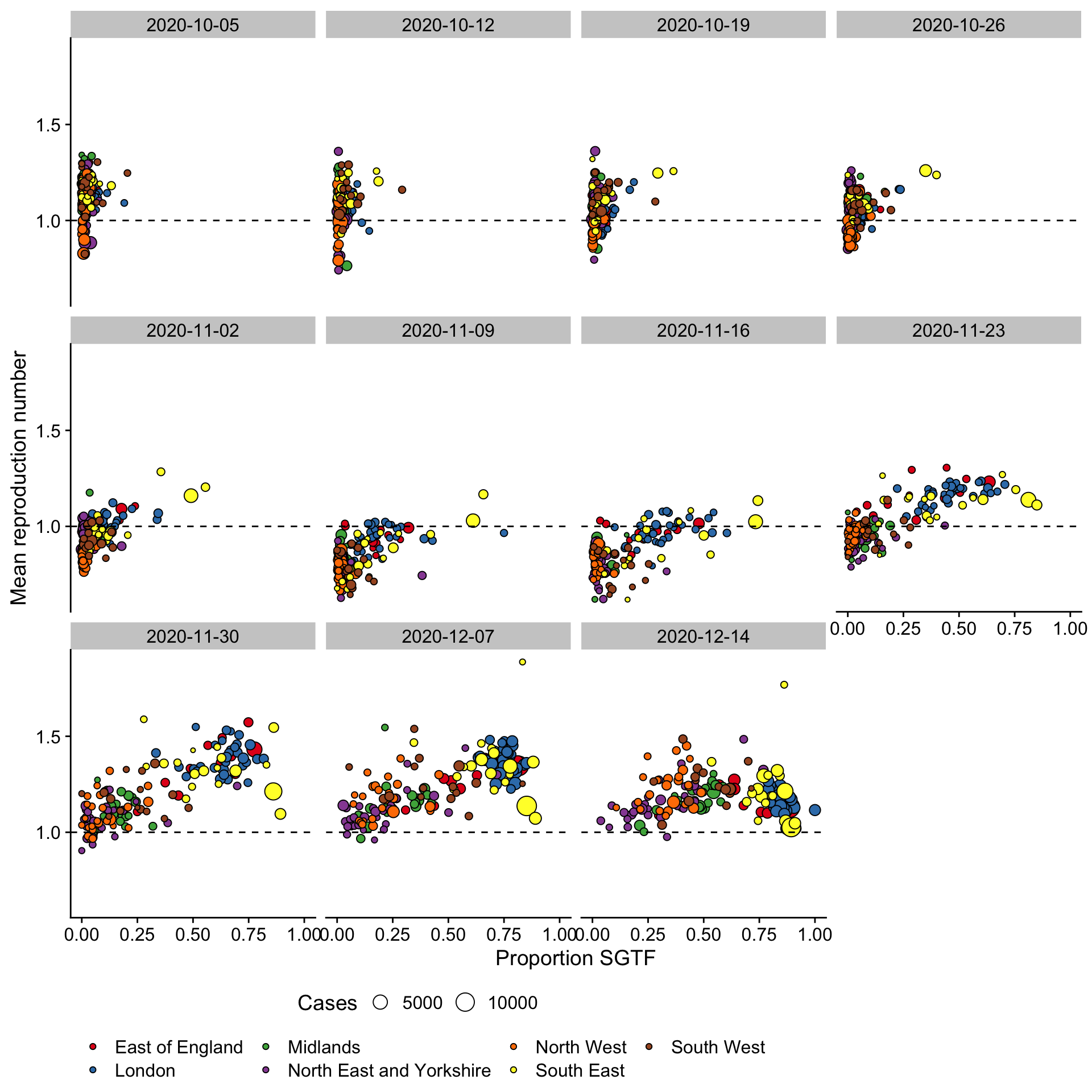
Figure 1: Proportion of test-positives that had a S-gene result reported which was S-gene positive/negative by LTLA and week of infection (assumed one week before test).